Research
Computational Biology
My research uses computational methods to analyze complex biological networks, and evaluates the computational methods with both computer simulation and biological intervention. By using computation and experimentation in concert, I can place a level of confidence on the results. I believe that computational biology should use practical methods that handle the inherent noise and error present in biological experiments, and that work using data that are practical to collect.
Read below about the research I perform in complex biological networks and my simulation framework for evaluating algorithms. Also visit my pages describing some background for the computational methods I work with:
Complex Biological Networks
My research concentrates on networks within three types of biological systems:
- Gene Regulation
- All our cells (with a few exceptions) have the same DNA, and yet cells in different tissues, and over developmental time, can have within them widely different proteins. This is achieved through differential gene regulation. When a gene is expressed, it is transcribed into mRNA, which is then translated into a protein product. Certain proteins, known as transcription factors, are responsible for regulating the expression of their target genes: either increasing/turning on (activation) or decreasing/turning off (suppression).A gene regulatory network is defined to be the network of genes which code for the transcription factor proteins, connected to their target genes. My research is concentrated on using and developing functional network inference algorithms to determine gene regulatory networks based on measurements of gene expression, such as from gene expression arrays. I use yeast, Saccharomyces cerevisiae, as a model system in which to develop and test these algorithms.Regulation also occurs at the step of protein translation as well as mRNA transcription, and thus the genes which code for proteins involved in this type of regulation also have a part in the gene regulatory network. I also focus on methods to incorporate this type of regulation into the network inference task.
- Neural Activity
- Neurons transmit electrical signals to one another by means of action potentials traveling down axons. The anatomical neural network consists of the physical connections between neurons, or collectively between entire brain regions, as defined by the paths of the axons. The neural information flow network is the subset of these connections which are in use during a certain behavioral state. While the anatomical neural network can be determined through physical examination, the neural information flow network can only be inferred from measurements of neural activity.My research is concerned with using and developing functional network inference algorithms to determine these neural information flow networks from measured activity. I am also interested in analysing the way the network changes for different behaviours and evolves over time throughout a behaviour.
- Ecological Systems
- Organisms in an ecological system depend upon each other for survival, are responsive to environmental conditions, and can even adjust the environmental conditions. Such relationships may be thought of as an ecological dependence network. My research involves using and developing functional network inference algorithms for determining ecological dependence networks based on species abundance and environmental data.
Simulation Framework for Evaluation of Algorithms
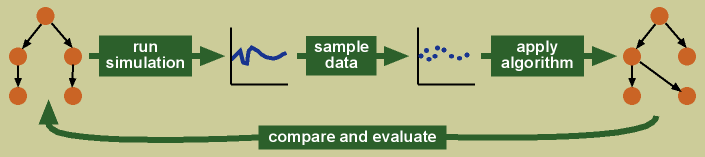
In many cases, the true network of the biological system to which we apply functional network inference algorithms is unknown (which is why we use the algorithms!). To biologically validate the networks produced by the algorithms is often infeasible—especially in the case of gene regulatory networks where hundreds to thousands of interactions may be predicted. To overcome this limitation, I have a developed a simulation framework for algorithm evaluation, which I introduced in the paper Evaluating functional network inference using simulations of complex biological systems (Smith et al. 2002 Bioinformatics 18:S216).
In my simulation framework, I create a simulation of a complex system, sample data from the running simulation as one would sample data in a biological experiment, and apply the algorithm to the sampled data. I then compare the networks predicted by the algorithm with the known truth underlying the simulation, and evaluate how well the algorithm has performed.
In addition to evaluating algorithm performance, my simulation framework also allows one to evaluate the effects of data collection and features of the underlying network on the outcome of the algorithm. The ability to examine effects of data collection on algorithm performance allows us to design biological experiments with computational analysis in mind. The ability to examine how the algorithm performs when given different underlying networks allows us to ask “what if?” questions about the algorithm’s ability. This is something that would be very difficult outside of a simulation framework, as even if one knew the underlying biology, it may not be possible to obtain such biological systems with the range of variation under consideration. The simulation framework allows us to evaluate the algorithm’s performance across a range of possibilities. See my papers Smith et al. 2003 PSB 8:164 and Yu et al. 2004Bioinformatics 20:3594 for these additional uses of my simulation framework.